 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
CSB Core Faculty
- Ray Blind
- Tony Capra
- Walter Chazin
- Steven Damo
- Jamaine Davis
- Martin Egli
- Brandt Eichman
- Steve Fesik
- Tina Iverson
- Lauren Jackson
- Erkan Karakas
- Brett Kroncke
- Borden Lacy
- Galina Lepesheva
- Carlos Lopez
- Terry Lybrand
- Hassane Mchaourab
- Jens Meiler
- Terunaga Nakagawa
- Yi Ren
- Chuck Sanders
- Ben Spiller
- Michael Stone
Emeritus:
- Al Beth
- Paul Bock
- Gerald Stubbs
- Michael Waterman
Jens Meiler
Structural and Chemical Biology

Research in our laboratory seeks to fuse computational and experimental efforts to investigate proteins, the fundamental molecules of biology, and their interactions with small molecule substrates, therapeutics, or probes. We develop computational methods with three major ambitions in mind: 1) to enable protein structure elucidation of membrane proteins - the primary target of most therapeutics - and large macromolecular complexes such as viruses; 2) design proteins with novel structure and/or function to explore novel approaches to protein therapeutics and deepen our understanding of protein folding pathways, and 3) understand the relation between chemical structure and biological activity quantitatively in order to design more efficient and more specific drugs. Crucial for our success is the experimental validation of our computational approaches which we pursue in our laboratory or in collaboration with other scientists. For a complete list of research projects please visit www.meilerlab.org. Current research projects include:
1) Protein Structure Elucidation of EMRE.
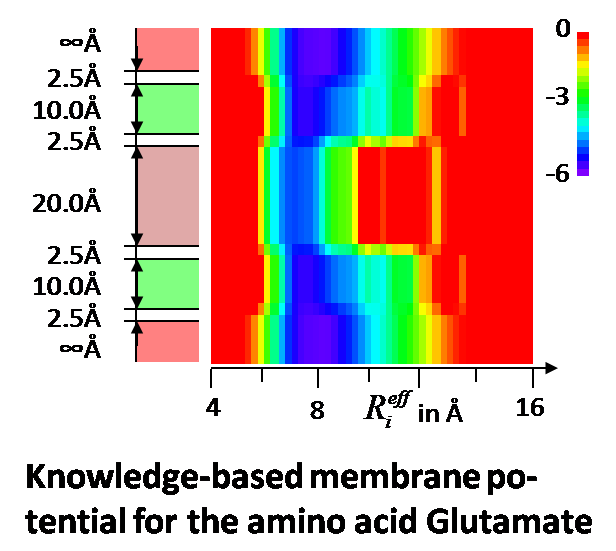 EmrE is a 12 kDa small multidrug resistant transporter (SMR) protein. It contributes to multidrug resistance in cancer and bacterial cells by removing compounds toxic to the cell such as the therapeutics. EmrE has been shown to contain four transmembrane alpha-helices and form a homodimer. While X-Ray crystallography and NMR spectroscopy frequently yield datasets for membrane proteins that are of lesser quality and/or sparse compared to soluble proteins, extensive Electron Paramagnetic Resonance (EPR) and cryo-Electron Microscopy (cryo-EM) datasets are available for EmrE. We develop computer algorithms tailored for determining the structure from these low resolution/sparse experimental data with the ultimate goal of solving the structure of EmrE and other membrane proteins. By determining the structure of EmrE, novel chemotherapeutic agents could be developed, including those to combat multidrug resistance.
EmrE is a 12 kDa small multidrug resistant transporter (SMR) protein. It contributes to multidrug resistance in cancer and bacterial cells by removing compounds toxic to the cell such as the therapeutics. EmrE has been shown to contain four transmembrane alpha-helices and form a homodimer. While X-Ray crystallography and NMR spectroscopy frequently yield datasets for membrane proteins that are of lesser quality and/or sparse compared to soluble proteins, extensive Electron Paramagnetic Resonance (EPR) and cryo-Electron Microscopy (cryo-EM) datasets are available for EmrE. We develop computer algorithms tailored for determining the structure from these low resolution/sparse experimental data with the ultimate goal of solving the structure of EmrE and other membrane proteins. By determining the structure of EmrE, novel chemotherapeutic agents could be developed, including those to combat multidrug resistance.
2) Design of Protein Antibiotics.
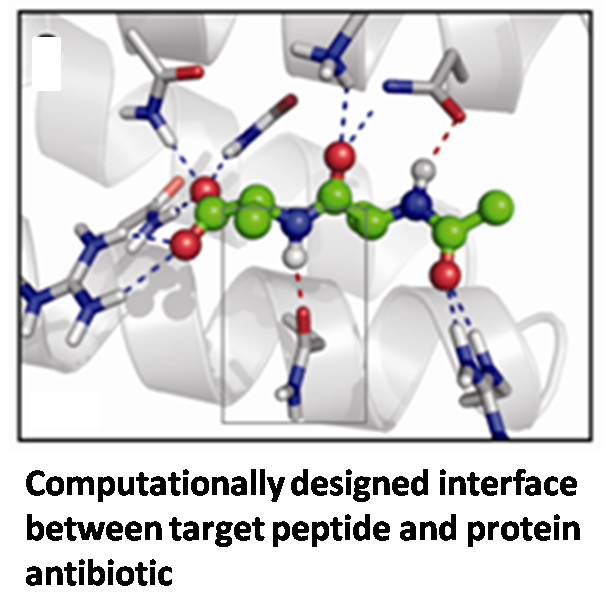 Gram positive bacterial infections are a significant global cause of human mortality. More than 125,000 people contract multidrug-resistant gram positive infections annually in the U.S. alone, resulting in more than 40,000 deaths per year. Vancomycin is the last-line antibiotic for gram-positive infections. It kills bacteria by binding the -D-ala-D-ala C-terminus of a key bacterial cell wall glycopeptide component, thereby inhibiting proper cell wall biosynthesis. The most common mechanism of acquired resistance is through the substitution of a -D-lac in place of the -D-ala at the C-terminus of the bacterial glycopeptide. The goal of this project is to explore a rational design approach to develop novel antimicrobial protein therapeutics capable of binding both the multidrug-resistant -D-ala-D-ala and vancomycin-resistant -D-ala-D-lac target peptides.
Gram positive bacterial infections are a significant global cause of human mortality. More than 125,000 people contract multidrug-resistant gram positive infections annually in the U.S. alone, resulting in more than 40,000 deaths per year. Vancomycin is the last-line antibiotic for gram-positive infections. It kills bacteria by binding the -D-ala-D-ala C-terminus of a key bacterial cell wall glycopeptide component, thereby inhibiting proper cell wall biosynthesis. The most common mechanism of acquired resistance is through the substitution of a -D-lac in place of the -D-ala at the C-terminus of the bacterial glycopeptide. The goal of this project is to explore a rational design approach to develop novel antimicrobial protein therapeutics capable of binding both the multidrug-resistant -D-ala-D-ala and vancomycin-resistant -D-ala-D-lac target peptides.
3) Novel Schizophrenia Therapeutics by Virtual High-Throughput Screening.
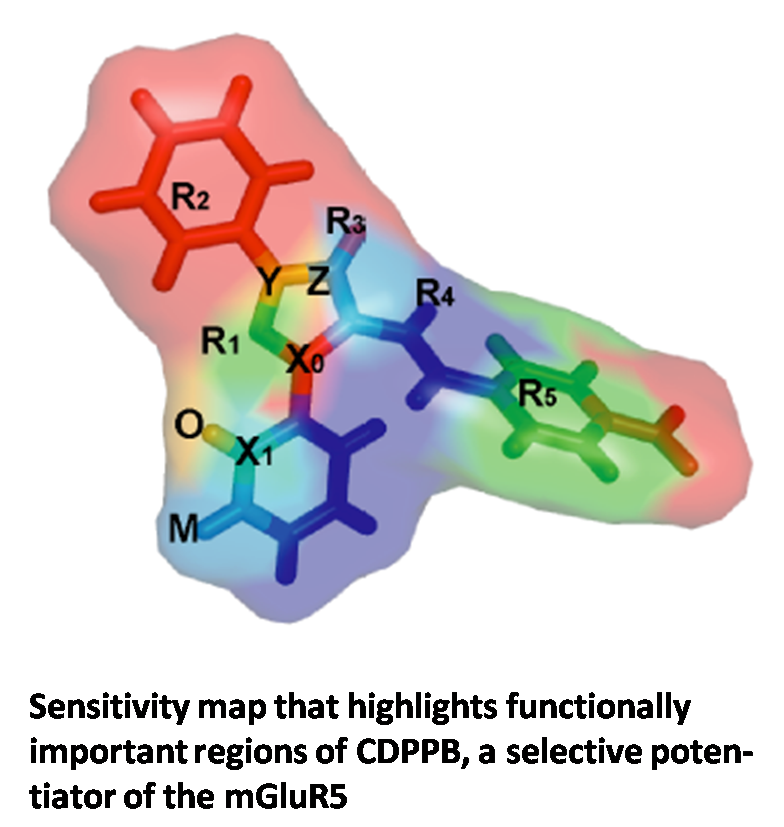 Selective potentiators of the metabotropic glutamate receptor subtype mGluR5 have exciting potential for development of novel treatment strategies for schizophrenia. A high-throughput screen (HTS) for mGluR5 potentiators at Vanderbilt's molecular libraries screening center network facility revealed a large and diverse set of about 1,400 substances. We utilize the power of recent machine learning techniques such as Artificial Neural Networks (ANNs) and Support Vector Machines (SVMs) to model the complex relationship between chemical structure and biological activity of mGluR5 potentiators. These models will be used to virtually screen millions of compounds for activity and guide chemical synthesis of novel compounds.
Selective potentiators of the metabotropic glutamate receptor subtype mGluR5 have exciting potential for development of novel treatment strategies for schizophrenia. A high-throughput screen (HTS) for mGluR5 potentiators at Vanderbilt's molecular libraries screening center network facility revealed a large and diverse set of about 1,400 substances. We utilize the power of recent machine learning techniques such as Artificial Neural Networks (ANNs) and Support Vector Machines (SVMs) to model the complex relationship between chemical structure and biological activity of mGluR5 potentiators. These models will be used to virtually screen millions of compounds for activity and guide chemical synthesis of novel compounds.
Jens is passionate about chemistry as the atomic resolution of the Chemist's view is crucial to address current challenges in biomedicinal research by linking physics governing the interaction between atoms and molecules to biological phenomena.
|
VU Home |
VUMC Home |
People Finder |
University Calendar
webmaster- modified on July 03, 2018 |