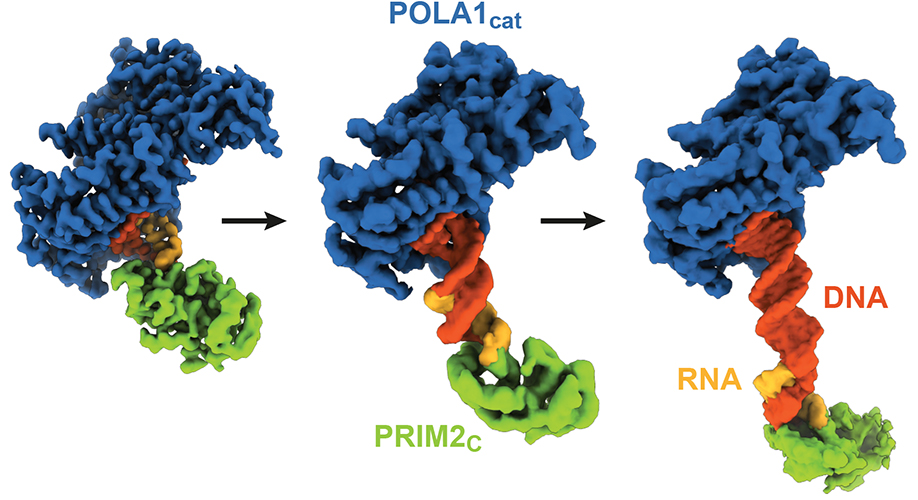
Cryo-EM structures of DNA polymerase α–primase during DNA synthesis
Read more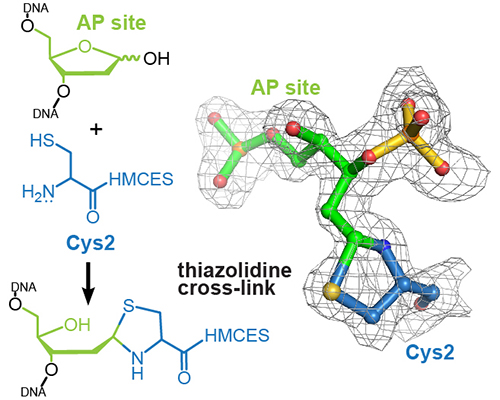
High-resolution structure of a native DNA-protein crosslink
Read more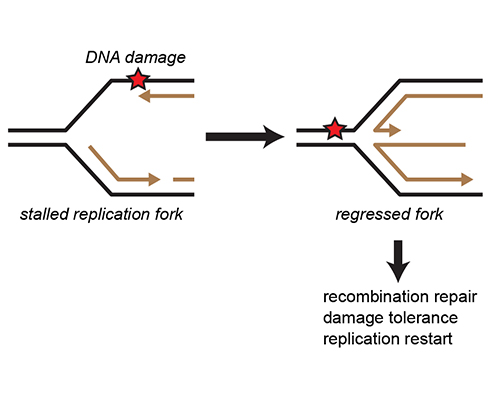
What is the mechanism of replication fork reversal?
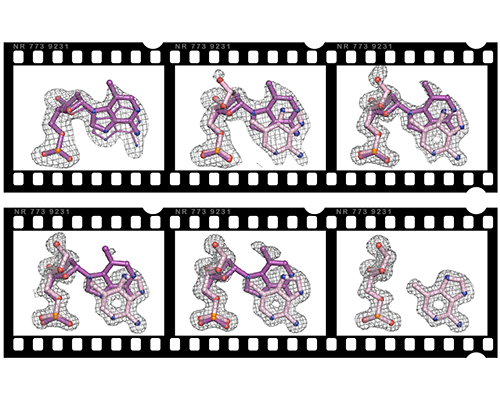
Time-resolved crystallography to monitor base excision repair by DNA glycosylases
Read more
HLTF's HIRAN domain binds 3' DNA ends to drive replication fork reversal
Read more
The DNA glycosylase YcaQ initiates an alternative interstrand DNA crosslink repair pathway in E. coli
Read more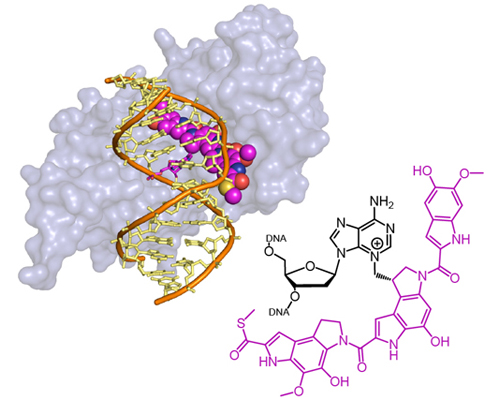
Bacterial AlkD repairs bulky DNA lesions formed by the natural product yatakemycin
Read more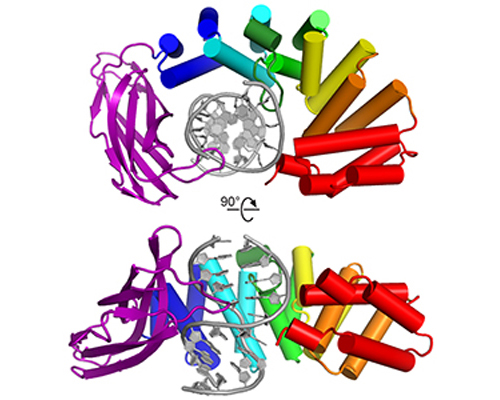
HEAT repeats have emerged as an important nucleic acid binding architecture
Read more
Reistance-based genome mining to identify new genotoxins
Read more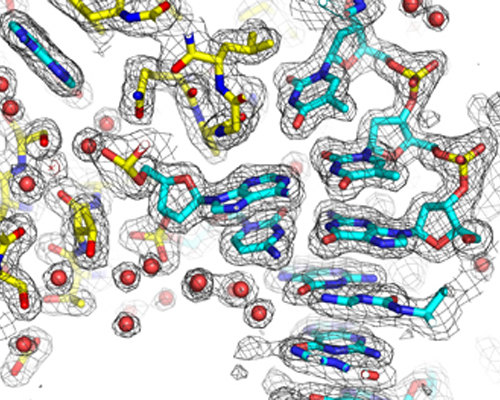
Building the atomic structure of a protein-DNA complex into experimental electron density from X-ray diffraction data
DNA Transactions at an Atomic Level
The genetic information encoded within DNA is copied, maintained, and decoded by protein machines. Our laboratory uses electron microscopy, X-ray crystallography, and other high-resolution structural and biochemical approaches to investigate the molecular details of how these proteins repair damaged DNA and maintain integrity of the genome during replication.
Featured Articles
An interstrand DNA crosslink glycosylase aids Acinetobacter baumannii pathogenesis. Proc Natl Acad Sci USA
In the News
Cassie Probert receives NSF Graduate Research Fellowship honorable mention
Leah Oswalt awarded NSF Graduate Research Fellowship
Article highlighting Elwood's work on polα–primase



