
Structural, biochemical, and functional studies of E3 Ubiquitin ligases
Signaling via protein ubiquitination involves the sequential activities of E1, E2 and E3 enzymes, which catalyze the activation and covalent attachment of ubiquitin to substrates. The E3 ligases have the task of recruiting both substrates and activated ubiquitin. A large number of proteins in the cell are ubiquitinated and there are correspondingly large numbers of E3 ligases with a diverse range of structures. The E3 ligases are broadly classified as simple (single polypeptide) and complex (multi-protein or multi-copy).
Our direct involvement in ubiquitination research was first prompted by a small structural project with a colleague, Kathy Gould (Department of Cell Biology, HHMI), which launched studies of the unique U-box family of E3 ligases. At about the same time, a primary cellular target of a calcium binding protein we had been studying for several years, S100A6, was reported to be part of the E3 ubiquitin ligase SCFTBL1. Once introduced to the complexities of ubiquitination machinery, I realized these are extremely fascinating multi-protein complexes for which structural and mechanistic studies would be highly useful.
1. U-box E3 Ligases
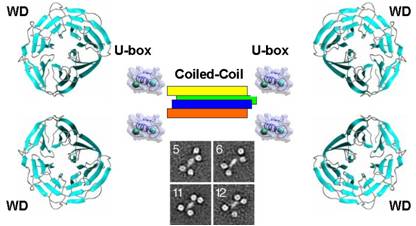
U-box proteins are a relatively new family of complex E3 ligases that are characterized by their unique U-box E2-recognition motif. The first U-box structure, from the yeast splicing factor Prp19 (Ohi 2003), revealed U-boxes have the same basic fold as the more prevalent RING domain E2-recognition motif. Structural studies ensued once Prp19 was shown to have ubiquitin ligase activity. We have constructed a model for the structure of the Prp19 tetramer using data from analytical ultra-centrifugation, negative stain cryo-electron microscopy and crystal structures of the U-box and substrate binding WD40 repeat domain (Vander Kooi 2009).
One of the continuing debates in the protein ubiquitination field is whether there are unique enzymes that specifically catalyze poly-ubiquitination, termed E4 ubiquitin ligases. The U-box E3 ligase CHIP is one protein purported to have E4 ligase activity, but only in combination with another U-box E3 ligase, E4B. Characterization of the structure, dynamics, and interactions of CHIP and E4B using the strategy developed for Prp19 is in progress, which should help determine if there are unique features of the complex that provide the putative E4 activity. We are also investigating the structural basis for the function of CHIP as an integral part of the 'protein triage' system; CHIP is able to target misfolded protein substrates for degradation via poly-ubiquitination as well as facilitate protein folding/refolding through stabilization of heat shock protein chaperone complexes. Current emphasis is on understanding the differential regulation by CHIP of the constitutive heat shock protein Hsc70 versus the stress-induced Hsp70 by determining differences in E2 selectivity, ubiquitination sites and chain linkages formed.
2. SCFTBL1 E3 Ligase
Initial interest in this complex E3 ligase was based on a connection to Ca2+ signaling (see below), then further motivated when its potential role in breast cancer was realized. The SCFTBL1 complex is assembled only under conditions of genotoxic stress when it is needed to poly-ubiquitinate -catenin to target the protein for proteasomal degradation. Elevated levels of -catenin are found in half of all breast cancers and directly correlate with low survival rates, so stimulation of the SCFTBL1 activity has become an interesting potential strategy for therapy. Our objective is to learn how the SCFTBL1 complex functions. Information on SCF E3 ligases is relatively scarce, so in addition to the specific interest in SCFTBL1, these studies should be of broader general interest. Protocols have been developed to produce each of the components of SCFTBL1 and for an in-vitro ubiquitination assay (Dimitrova 2009). Our major goal would be to determine the mechanism of -catenin recruitment and poly-ubiquitination by the SCFTBL1 E3 ligase utilizing a range of structural, biochemical and functional techniques.
- Understanding the mechanics of multi-protein DNA processing machinery
- Structural, Biochemical, and Functional Studies of E3 Ubiquitin Ligases
- Ca2+ Signal Trasnduction by EF-hand Proteins