
Understanding the mechanics of multi-protein DNA processing machinery
Fundamental processes in biology such as DNA replication and repair require the coordinated activity of a number of proteins because they involve multiple steps of molecular recognition and chemistry. I am interested in investigating the dynamic nature of multi-protein complexes and the transitions that need to occur for forward progression of these multi-protein machines. To this end, we have starting using solution NMR and scattering approaches to complement the snapshots of multi-protein complexes that can be obtained by X-ray crystallography and cryo-electron microscopy. The objective in this research is to understand how each protein works as part of the corresponding complex. We are developing an understanding of the common principles that relate the modular nature of these proteins and the multiple contact points between binding partners to structural mechanism and function (e.g. Stauffer 2004).
A strategic decision was made to focus initial efforts on Replication Protein A (RPA), the primary single-strand DNA (ssDNA) binding protein in eukaryotic cells. RPA is a required factor for virtually all DNA transactions, and essential functional interactions have been identified with a wide range of proteins. Indeed, it is now well accepted that RPA functions in a highly interwoven series of steps involving binding of ssDNA, internal structural rearrangements, and interactions with other proteins. The initial work from our laboratory (Mer 2000) revealed RPA uses a common binding mode for DNA damage recognition proteins from three different DNA repair pathways. These results suggested that RPA serves as a scaffold for the ordered assembly and disassembly of DNA processing machines.
1. SV40 DNA replication
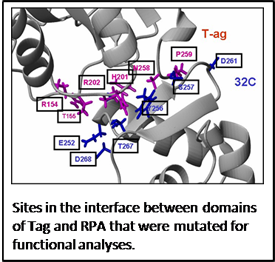
To develop a comprehensive view of RPA function, it is important to examine all of its roles in a single system. I selected DNA replication initiated by Simian Virus 40 (SV40) for these efforts because it is a simple and very well studied system. Remarkably, a single protein from SV40, the large T antigen (Tag), is able to initiate all of the earliest stages of DNA replication using the RPA and DNA polymerase /primase (DNA pol-prim) from the host organism. We are systematically studying this coupled set of proteins beginning with biochemically and structurally mapping out the network of binding sites. The interactions are then quantified using biophysical methods and the interfaces are characterized structurally using a combination of crystallography, NMR and computational docking. The information obtained is used in turn to design mutations for functional studies in-vitro and in-vivo in collaboration with Ellen Fanning (Dept. of Biological Sciences). The first two papers using this approach (Arunkumar 2005, Jiang 2006) demonstrate how fundamental insights into the mechanisms for the coordinated action of these proteins can be obtained.
2. RPA Quaternary Structure
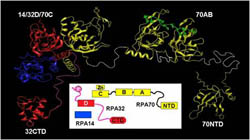
Knowledge of RPA quaternary structure and remodeling by DNA and RPA-binding proteins is essential to understanding RPA function. RPA represents a significant challenge because it is a heterotrimer of 70 kDa (yellow), 32 kDa (red) and 14 kDa (blue) subunits that contain seven different structural domains attached by linkers of varying length and flexibility. Despite the fact that highly modular proteins like RPA are quite common, knowledge of the quaternary structure and of changes in the architecture of such proteins is extremely scarce. A critical aspect for studying RPA is that high resolution structures are available for all seven globular domains, which enables focusing directly on quaternary structure. The approach being developed involves NMR heteronuclear relaxation and residual dipolar coupling experiments in combination with X-ray scattering to characterize the flexibility of the linkers and the dynamic spatial organization of the domains (Brosey 2009, Pretto 2009). We are also examining the effects of poly-phosphorylation on the structure and dynamics of RPA to understand the basis for this critical post-translational modification, which serves to promote the switch from replication to repair processing of DNA.
- Understanding the mechanics of multi-protein DNA processing machinery
- Structural, Biochemical, and Functional Studies of E3 Ubiquitin Ligases
- Ca2+ Signal Trasnduction by EF-hand Proteins