
Wisdom Page: Kd
measurement, fitting, calculation, and simulation
This wisdom page covers Kd measurements and calculations in direct
measurement mode (the type of data that would be collected by fluorescence or
NMR, for example). Similar
considerations apply for data collected in the derivative mode (ITC data), not
discussed here. I only treat single site
binding here; this will cover 98% of the applications we will encounter in the Chazin lab. Again,
for multiple site binding the considerations are similar but (obviously)
somewhat more complex.
Deriving and simulating the equations governing the behaviour of protein/ligand
interactions can provide a significant base for understanding the limits of this
measurement, including optimal concentration ranges for measuring certain
ranges of Kd. Critically, these simulations can provide an
absolute floor or ceiling on the amount of error that can be expected in a
given calculation.
To begin at the beginning:
If protein P binds ligand A, then an equilibrium is formed:
P + A < -- > PA
Where P and A are free protein; PA
is the ligand-bound-protein.
There is a ratio between the free species and the bound
species.
At a given set of conditions, this ratio is fixed:
Sometimes [P] is called [Pfree], and likewise [A] = [Afree]. Substituting, we get:
Now remember that the total amount of protein is divided into two "camps," free protein and ligand-bound-protein. Likewise for the total amount of ligand:
[Ptotal] = [Pfree] + [PA] which can be rewritten as [Pfree] = [Ptotal] - [PA]
[Atotal] = [Afree] + [PA] which can be rewritten as [Afree] = [Atotal] - [PA]
Therefore:
![]()
A little algebra as follows:
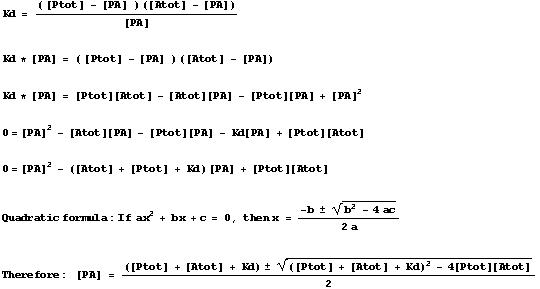
Immediately below is an important but simple equation that represents our assumption that the response being measured (Q) varies linearly with the proportion of protein in the bound state as compared to total protein present. Combining this equation with our results from the quadratic equation above yields the final equation.
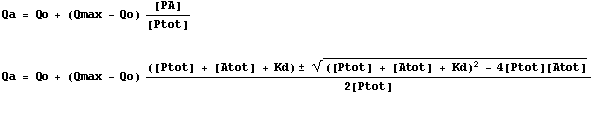
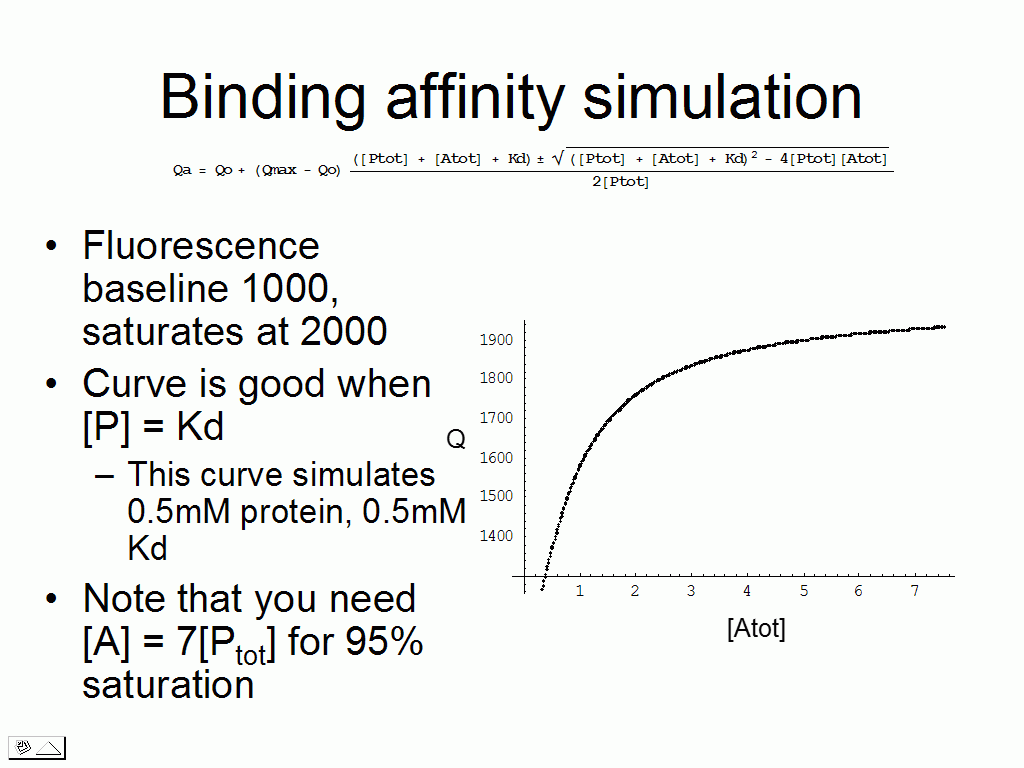
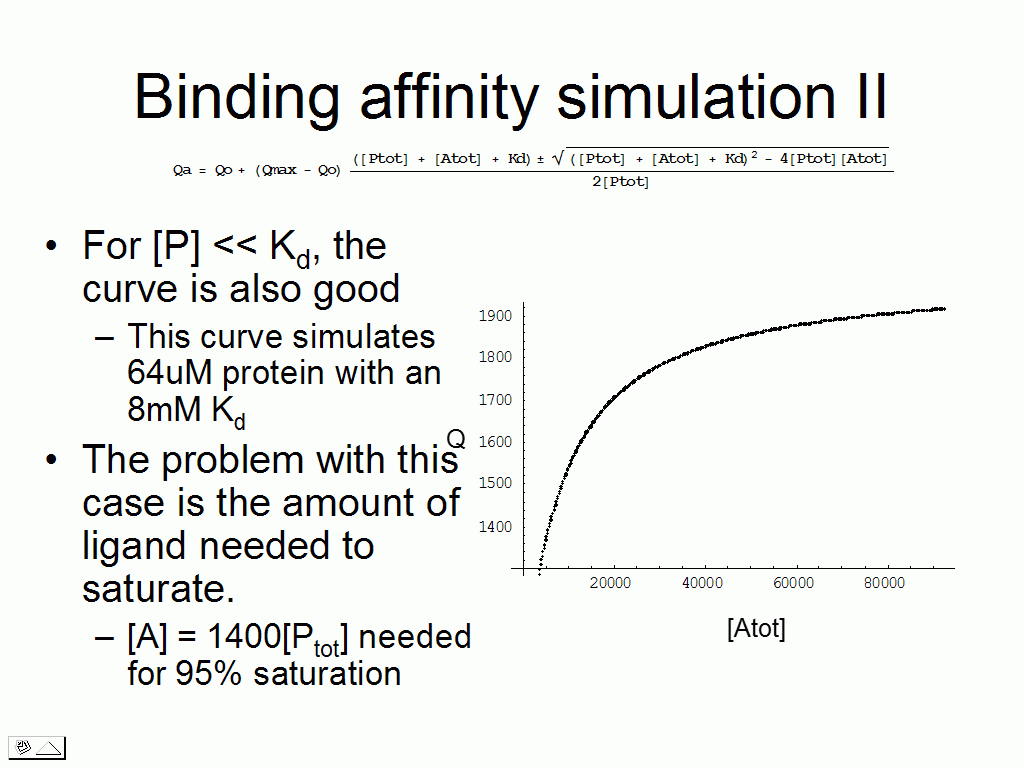
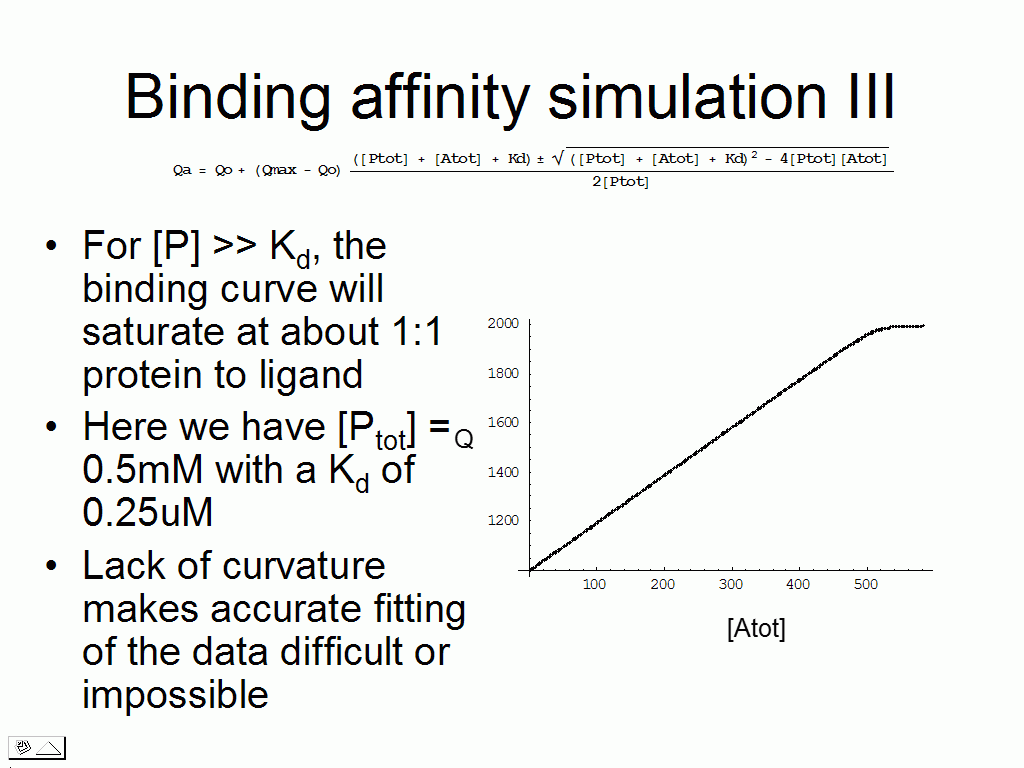
We can test various scenarios to realize that for certain
combinations of Kd and
protein concentration, there are issues to making the measurement (requires a
vast excess of ligand, the binding plot is linear).
Below I provide a Mathematica
program for running these simulations.
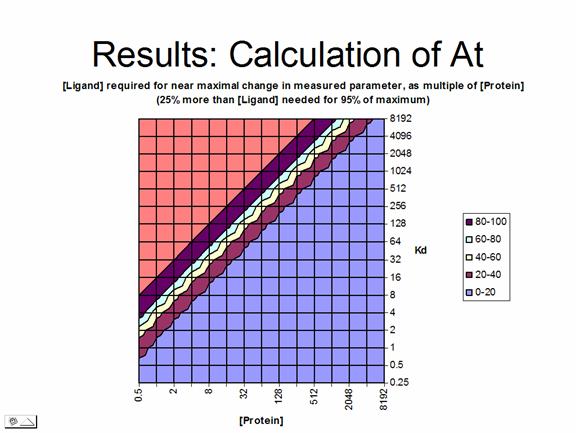
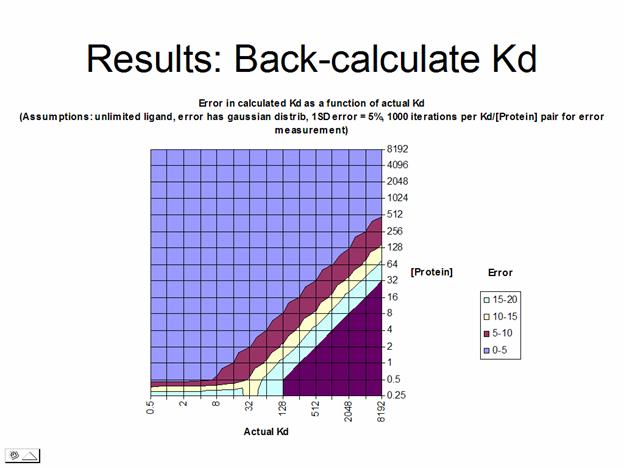
The overall results of the calculation testing a wide range of Kd/protein concentration combinations is that protein in excess of 10-50*Kd results in very large errors in measurement, whereas small amounts of protein with weak binding require a large excess of ligand to achieve saturation. These results are summed up in the two graphs beow.
Instructions on using the Mathematica
program for Kd simulation:
To get the Mathematica notebook, right click the above link and "Save Target As" to a folder. The notebook was written and tested in Mathematica 4.0.
Enter the parameters.
The first two parameters are self descriptive. The
program assumes that the units used for [Protein] and Kd are IDENTICAL.
Therefore if you estimate ~10uM Kd
but have 0.5mM protein, adjust one of the numbers so that the units match.
'EstimatedPercentSaturation' tells
the program how far out it should go in the simulation. Do NOT enter values at above 100, the program will run
forever.
'PercentGaussianError' tells the
program how much error to introduce at each data point. The percent given by here will represent 1
standard deviation in a Gaussian distribution around the value of the data
point. While this leads to a violation
of one of the fundamental assumptions
of nonlinear regression, the effect
is minor.
'IterationsForErrorEvaluation'
controls the number of times that the program should run with the combination
of parameters you've entered. On each
iteration, the program compares the back -calculated Kd to the actual starting Kd
to achieve an absolute value error estimate.
Once you've entered your parameters, press [SHIFT] - [ENTER]
to run the program.
Output consists of a representative plot of the simulated binding
titration with and without Gaussian error introduced. Three numbers follow:
the
first is the error in Kd calculated over the number
of iterations specified,
while
the second and third are the overall average values obtained for fitting Qo and Qmax.
This is followed by a
representative set of fitting diagnostics from a single iteration of the
nonlinear fit. This includes standard
error, confidence
intervals, etc. The final number
displayed is the number of times the calculation
failed.