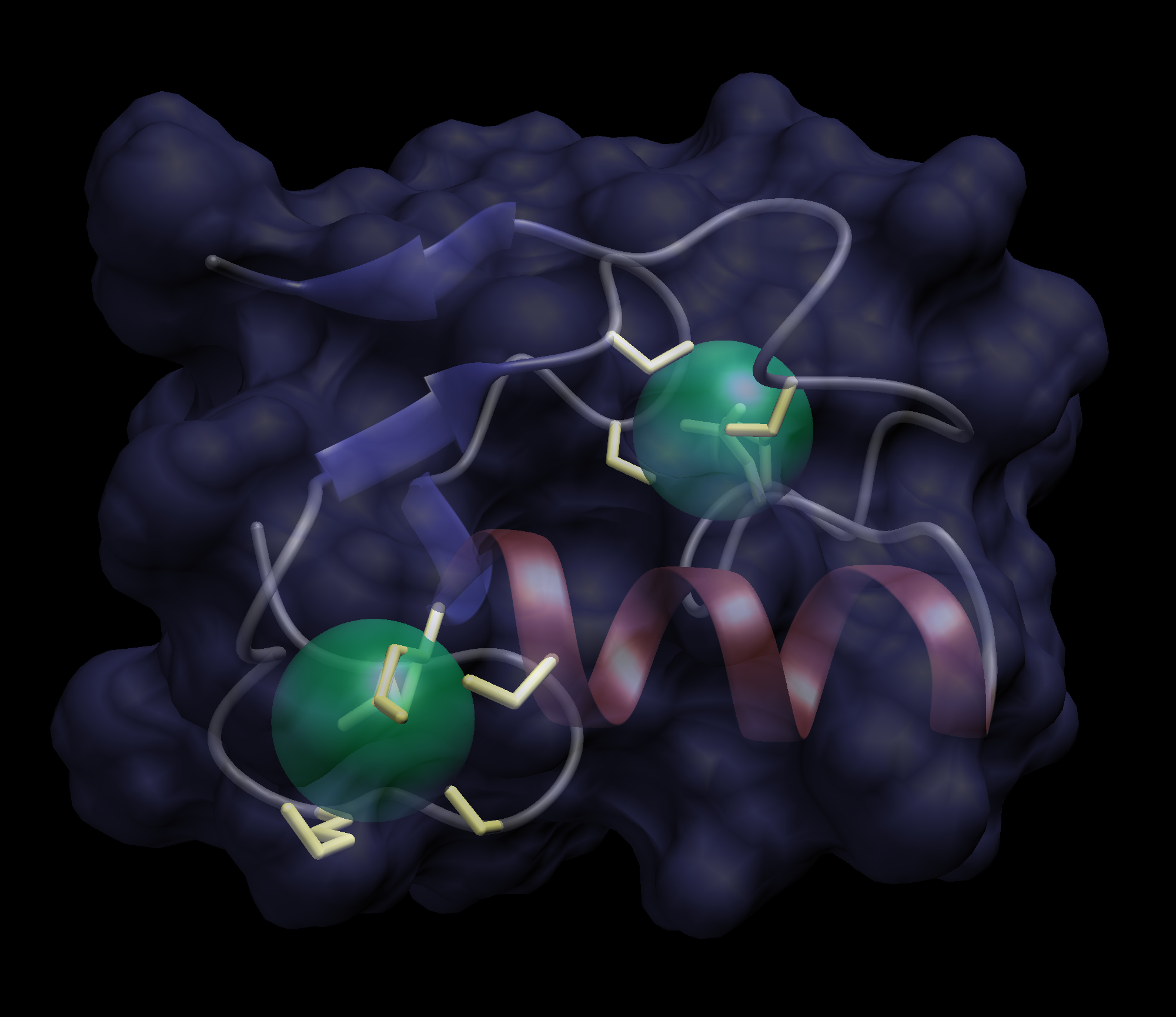
This is a tutorial to make cool Dino renderings of proteins
Features included
Protein surface creation and transparent display
BB and select side chain display
Variable colors on all
Let me know if you want to include additional features
Start with your PDB file, for instance protein123.pdb
1) Convert to xyz coordinate file
>pdb_to_xyzrn protein123.pdb > protein123.xyzrn
2) Create msms surface (density determines quality of surface created)
>msms -if protein123.xyzrn -of protein123 -density 4.0
3) Modify your dino scr file.
Just a note, colors can be in two forms
1) color=$name (name=white, black, blue, light blue...)
2) color={r,g,b} (red,green,blue ex {.8,.3,.3})
Here is a commented example.
// Define some variables with the HELIX and SHEET records
// from the PDB file (columns 5and7 for HELIX, 6and8 for sheet)
// Or, use MolMol calcsecondary, save to text and fix up...
set helix rnum=402:410
set strand rnum=391:393,398:400,426:428
// Load the pdb file
load protein123.pdb
// Set the secondary structure based on the variables
// we defined above
.protein123 set rtype=helix -sel $helix
.protein123 set rtype=strand -sel $strand
// Draw the molecule (stick figure) and call it 'all'
.protein123 new -name all
// Center the molecule, then hide it
scene center [.protein123.all]
.protein123.all hide
// Create a C-alpha trace and define our helices and sheets
.protein123 new -type trace -name ca
.protein123 set rtype=helix -sel $helix
.protein123 set rtype=strand -sel $strand
// Render the secondary structure as a ribbon
// You can use customize the width and thickness, too
.protein123.ca render hsc,strandw=.8,strandt=0.15,helixt=0.15,helixw=0.8,arrowt=0.6,detail=20
// Set the color of the ribbon
.protein123.ca set color=white
.protein123.ca set color=white -sel $helix
.protein123.ca set color=white -sel $strand
// Render with the new color in effect and transparency t (in %)
.protein123.ca render t=0.3
// Change the specular highlighting (shininess) (Two lights used in this
// example. Lighting is very important, play with it!)
scene:light0 set spec=2.0,diff=0.6,pos={50,100,5,1}
scene:light1 set pos={1,2,5,1}, local, spec=3.0,diff=0.6
scene:light1 on
// Customize the "custom" rendering
.protein123.sc render custom,bw=.2,sr=2.0,detail=10
// white background, less fog
// scene set bg=white
// scene set fogd=1.0
// Load and display the surface with color {r,g,b} and transparency t
load protein123 -type msms -name surf
.surf attach .protein123
.surf new -name all
.surf.all set color={0.2,0.2,0.6}
.surf.all render t=0.3
// TURN ON INTERESTING SIDECHAINS
// Restrict so that backbone atoms aren't displayed
.protein123 restrict not aname=C,O,N,H*
// Make sidechain selection
set sidechains rnum=3,6,8,24,26,16,19,38,41,46
.protein123 new -name sc -sel $sidechains
// Color the sidechains
.protein123.sc set color={1,1,.5}
.protein123.sc render custom,sr=0.2,bw=0.2
4) In order to get transparency working correctly you need to use dino.test
>dino.test -s protein123.scr
5) Print to a graphics file using "-s res"
write protein123.png -s 2000
You may also output to povray, ps, tif, and others
Here is one example that I constructed. Let me know if you have any questions, cvanderk@
(click for large version)