 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
AMBER Archive (2008)
Subject: Re: AMBER: 3fe-4s cluster parameter file generation
From: moitrayee_at_mbu.iisc.ernet.in
Date: Tue Sep 16 2008 - 06:39:25 CDT
- Next message: chaitanya koppisetty: "AMBER: nmode/nab entropy calculations memory issues"
- Previous message: Waqas Nasir: "Re: AMBER: top and crd files without hydrogen"
- In reply to: Bill Ross: "Re: AMBER: 3fe-4s cluster parameter file generation"
- Next in thread: Bill Ross: "Re: AMBER: 3fe-4s cluster parameter file generation"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
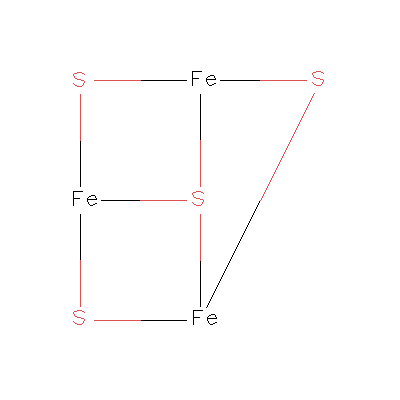
I am attaching the structure of my ligand. In that case is my prepc file
connectivities alright.
Looking eagerly forward to hear from you
Sincere Regards
Moitrayee
>> 1 DUMM DU M 999.000 999.0 -999.0 .00000
>> 2 DUMM DU M 999.000 -999.0 999.0 .00000
>> 3 DUMM DU M -999.000 999.0 999.0 .00000
>> 4 S1 S S 0.402592 -1.514288 -1.117435 -0.15436
>> 5 S2 S S -1.882611 0.670231 -0.727350 -0.17413
>> 6 S3 S B 0.078709 -0.205500 1.731896 -0.31438
>> 7 S4 S S 1.046182 1.057400 -1.309825 -0.13664
>> 8 FE1 FE B -1.129336 -1.094156 0.156620 0.63632
>> 9 FE2 FE B 1.553155 -0.302094 0.294594 0.59276
>> 10 FE3 FE B -0.205279 1.391423 0.424303 0.55042
>
> Looking at the doc of the prep format:
>
> http://amber.scripps.edu/doc/prep.html
>
> I think you have something like
>
> S-S-S-S-Fe
> |
> Fe-Fe
>
> What bonds are you trying to create? Maybe something like this:
>
> S-S-S-S-Fe
> | |
> Fe Fe
>
> In which case the topological types might be:
>
> M-M-M-M-E
> | |
> E E
>
> And the order of atoms:
>
> 1-2-4-6-7
> | |
> 3 5
>
> This may help you think out your structure in any case.
>
> Bill
> -----------------------------------------------------------------------
> The AMBER Mail Reflector
> To post, send mail to amber_at_scripps.edu
> To unsubscribe, send "unsubscribe amber" (in the *body* of the email)
> to majordomo_at_scripps.edu
>
> --
> This message has been scanned for viruses and
> dangerous content by MailScanner, and is
> believed to be clean.
>
>
-- This message has been scanned for viruses and dangerous content by MailScanner, and is believed to be clean.
----------------------------------------------------------------------- The AMBER Mail Reflector To post, send mail to amber_at_scripps.edu To unsubscribe, send "unsubscribe amber" (in the *body* of the email) to majordomo_at_scripps.edu
- Next message: chaitanya koppisetty: "AMBER: nmode/nab entropy calculations memory issues"
- Previous message: Waqas Nasir: "Re: AMBER: top and crd files without hydrogen"
- In reply to: Bill Ross: "Re: AMBER: 3fe-4s cluster parameter file generation"
- Next in thread: Bill Ross: "Re: AMBER: 3fe-4s cluster parameter file generation"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
|
VU Home |
VUMC Home |
People Finder |
University Calendar
webmaster- modified on January 30, 2009 |