 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
AMBER Archive (2008)
Subject: AMBER: trouble interpreting <P2> time correlation function data
From: Sally Pias (sallypias_at_gmail.com)
Date: Sun Jul 06 2008 - 01:59:57 CDT
- Next message: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Previous message: Ross Walker: "RE: RE: AMBER: about QMMM output"
- Next in thread: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Reply: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
I have run long MD simulations of a protein in explicit solvent (using
NMR distance restraints) and am attempting to calculate the order
parameter S2 for comparison with experimental data. I have, thus, used
ptraj to calculate auto-correlation functions, <P2>, from which I
hoped to determine S2 values (as the long-time asymptote of <P2>).
However, I have found the <P2> data to be confusing.
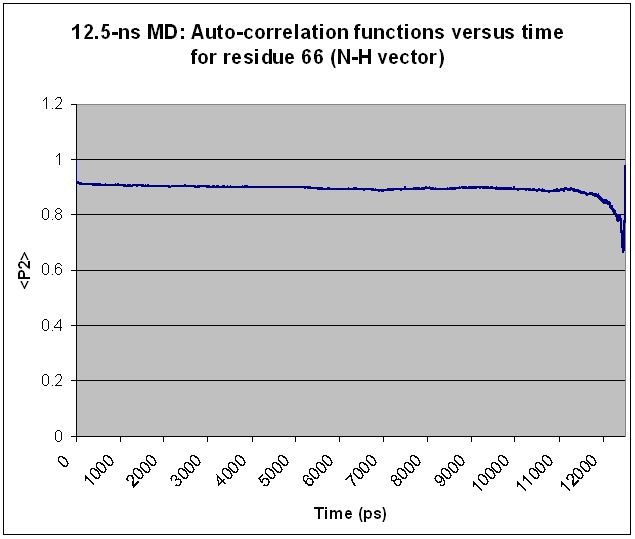
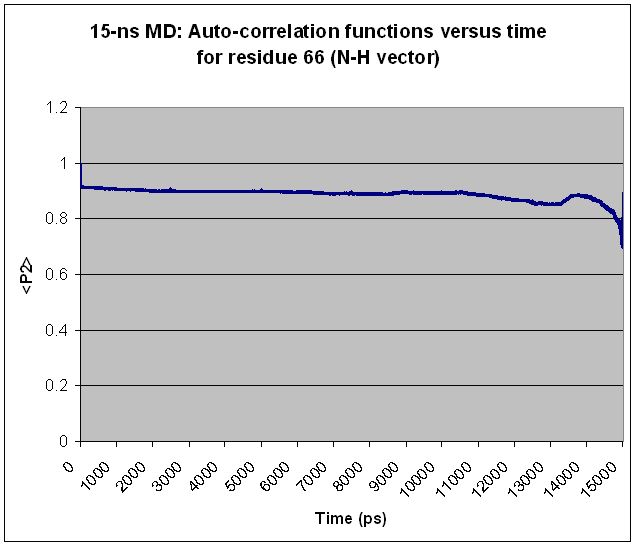
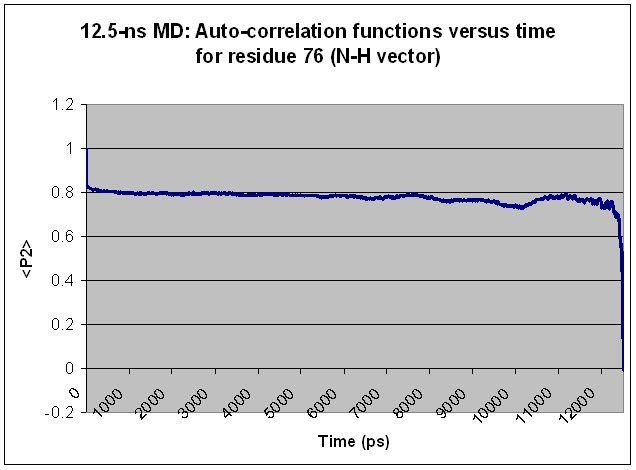
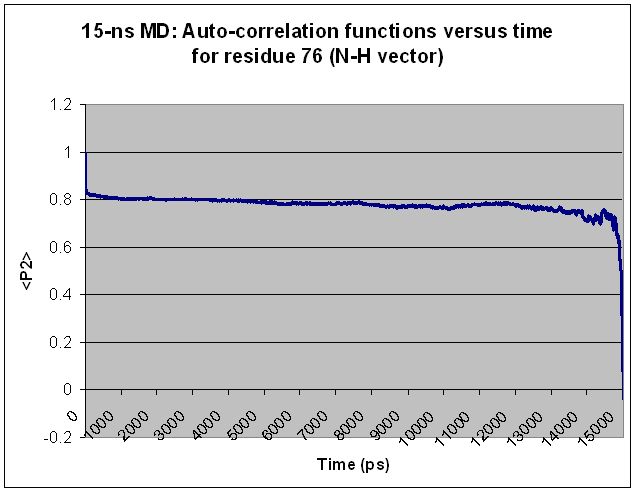
Specifically, I find that every residue shows "volatility" in <P2>
toward the end of the simulation, regardless of simulation length and
regardless of the residue's relative mobility. Extending the
simulation merely shifts the volatile region over but does not change
the basic shape of the <P2> curve. For most residues, what appears to
be a volatile region in a shorter simulation becomes smooth as the
simulation is extended. Two examples are given below, along with the
input file used to generate the <P2> values for the longer simulation.
(Graphs are attached showing <P2> values calculated from 12.5-ns and
15-ns simulations for each of two different residues.)
ptraj input file for the 15-ns simulation:
=====
trajin ../wtligand-mod6_heat.mdcrd.gz
trajin ../wtligand-mod6_1st2500ps-rst.mdcrd.gz
trajin ../wtligand-mod6_2nd5000ps-rst.mdcrd.gz
trajin ../wtligand-mod6_3rd2500ps-rst.mdcrd.gz
trajin ../wtligand-mod6_4th2500ps-rst.mdcrd.gz
trajin ../wtligand-mod6_5th2500ps-rst.mdcrd.gz
rms first out wtligand-mod6_0-15020ps_core_op.rms :17-116_at_CA,C,N,O time 0.5
vector r2 :2_at_N corr :2_at_H
vector r4 :4_at_N corr :4_at_H
vector r5 :5_at_N corr :5_at_H
vector r6 :6_at_N corr :6_at_H
vector r7 :7_at_N corr :7_at_H
vector r8 :8_at_N corr :8_at_H
vector r9 :9_at_N corr :9_at_H
vector r10 :10_at_N corr :10_at_H
vector r11 :11_at_N corr :11_at_H
vector r12 :12_at_N corr :12_at_H
.
.
.
vector r127 :127_at_N corr :127_at_H
vector r128 :128_at_N corr :128_at_H
vector r130 :130_at_N corr :130_at_H
analyze timecorr vec1 r2 vec2 r2 out r002.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r4 vec2 r4 out r004.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r5 vec2 r5 out r005.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r6 vec2 r6 out r006.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r7 vec2 r7 out r007.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r8 vec2 r8 out r008.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r9 vec2 r9 out r009.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r10 vec2 r10 out r010.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r11 vec2 r11 out r011.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r12 vec2 r12 out r012.nh tcorr 15020. tstep 0.5
.
.
.
analyze timecorr vec1 r127 vec2 r127 out r127.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r128 vec2 r128 out r128.nh tcorr 15020. tstep 0.5
analyze timecorr vec1 r130 vec2 r130 out r130.nh tcorr 15020. tstep 0.5
go
 -----------------------------------------------------------------------
-----------------------------------------------------------------------
The AMBER Mail Reflector
To post, send mail to amber_at_scripps.edu
To unsubscribe, send "unsubscribe amber" (in the *body* of the email)
to majordomo_at_scripps.edu
- Next message: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Previous message: Ross Walker: "RE: RE: AMBER: about QMMM output"
- Next in thread: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Reply: David A. Case: "Re: AMBER: trouble interpreting <P2> time correlation function data"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
|
VU Home |
VUMC Home |
People Finder |
University Calendar
webmaster- modified on January 30, 2009 |