 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
AMBER Archive (2007)
Subject: AMBER: Membrane Simulation: NPT Issues
From: Akshay Patny (akshay17_at_olemiss.edu)
Date: Wed Feb 07 2007 - 11:28:05 CST
- Next message: WANG,YING: "Re: AMBER: Help!! I cannot perform the ambpdb"
- Previous message: Rima Chaudhuri: "AMBER: Binding free energy calculation - MM-pbsa"
- Next in thread: Akshay Patny: "RE: AMBER: Membrane Simulation: NPT Issues"
- Reply: Akshay Patny: "RE: AMBER: Membrane Simulation: NPT Issues"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
Dear Amberites
I am doing a GPCR membrane simulation.
After stepwise minimization of my system, I did a 20ps NVT simulation from
0-100 K followed by an NPT simulation of 100 ps from 100-300K.
Everything looked okay until 20ps NVT simulation (to 100K). However, when I
applied NPT conditions and raised temp to 300K, the inner/outer water layers
got displaced.
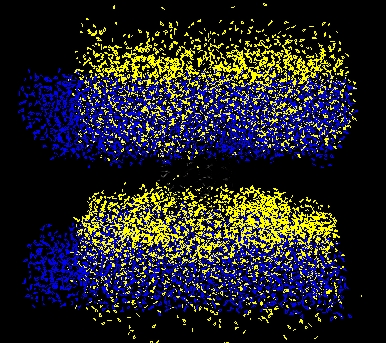
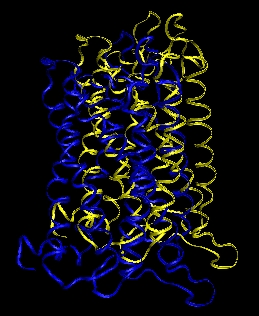
I am attaching 2 pictures here (water and protein):
BLUE: After 20 ps of NVT
YELLOW: After additional 100ps of NPT
My last 100ps NPT input file is as follows:
----------------------------------------------------------------------------
----------------------------------------------------------------------------
------------------------------------------------
Angi Apo in DMPC Water Ions: Second 100ps NPT Equilibration MD with harmonic
rest on Protein backbone: Heat from 100-300 K
&cntrl
imin = 0,
irest = 1,
ntx = 7,
ntb = 2,
ntp = 2,
pres0 = 1.0,
taup = 2.0,
cut = 10,
ntr = 1,
ntc = 2,
ntf = 2,
tempi = 100.0,
temp0 = 300.0,
ntt = 3,
gamma_ln = 1.0,
nstlim = 50000, dt = 0.002,
ntpr = 100, ntwx = 100, ntwr = 1000
restraint_wt=10.0
restraintmask=':1-297_at_CA,C,O,N,H'
/
----------------------------------------------------------------------------
----------------------------------------------------------------------------
------------------------------------------------
My questions are:
Q.1 Is this displacement of water,protein (system) normal? I mean I
understand that applying NPT conditions will adjust the volume of cell to
increase or decrease?
Q.2 I am using ntb = 2, ntp = 2, pres0 = 1.0, taup = 2.0
2.A. Since I am simulating a membrane protein, and my BOX is RECTILINEAR (as
mentioned in OUT file), I considered using ntp = 2 which is anisotropic
pressure scaling? I want to confirm if it makes sense or shall I use ntp =
1?
2.B. In addition, I used taup = 2.0. In the mannual it says that deafult
value is 1.0 but higher values might be necessary if trajectories are
unstable. What should be the reasonable value of this parameter to be used
in membrane protein simulation.
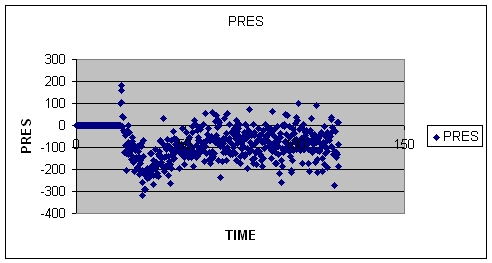
2.C. NOTE: I am also attaching a pressure/time picture which also suggests
that during last 100 ps, pressure has not be stabalized around 1 atm. What
can I do to to take care of this issue?
Q.3. Is harmonic restraint of 10.0 too low, shall I increase it to 50.0 or
something?
I will be great help if you can provide me some suggestions to solve these
problems.
Thanks and Best Regards
Akshay
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
Akshay Patny
PhD Candidate (5th Yr.), Computational Chemistry
Department of Medicinal Chemistry, School of Pharmacy
The University of Mississippi
805 College Hill Rd, # 9, Oxford, MS 38655
E-mail: akshay17_at_olemiss.edu
Phone (O): (662)-915-1286,(M): (662)-801-5496
= = = = = = = = = = = = = = = = = = = = = = = = = = = = = =
-----------------------------------------------------------------------
The AMBER Mail Reflector
To post, send mail to amber_at_scripps.edu
To unsubscribe, send "unsubscribe amber" to majordomo_at_scripps.edu
- Next message: WANG,YING: "Re: AMBER: Help!! I cannot perform the ambpdb"
- Previous message: Rima Chaudhuri: "AMBER: Binding free energy calculation - MM-pbsa"
- Next in thread: Akshay Patny: "RE: AMBER: Membrane Simulation: NPT Issues"
- Reply: Akshay Patny: "RE: AMBER: Membrane Simulation: NPT Issues"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
|
VU Home |
VUMC Home |
People Finder |
University Calendar
webmaster- modified on January 30, 2009 |