 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
CSB Biomolecular Visualization Workshop - July 2010
The Center for Structural Biology will teach a workshop in Biomolecular Visualization. This is an introductory workshop and assumes no prior knowledge or experience in the areas of molecular modeling or visualization. Beginners are encouraged to participate, as this is the target audience for this workshop.Enrollment info
- This workshop is offered to all Vanderbilt students/faculty/staff. The fee for this six-hour workshop is $75.
- Advance registration is required due to limited capacity in the molecular modeling teaching lab. Enrollment is limited to 13 people on a first-come, first-served basis. The class has filled quickly in the past. Therefore, register early to ensure the availability of your seat.
Workshop Syllabus
The six-hour workshop will be held July 7th - 8th, 2010 in the College of Arts and Sciences Molecular Modeling Teaching Laboratory (5119 Stevenson Center) from 1-4pm each day. There are approximately two hours of lecture and four hours of hands-on work at the computer, with assistance from the instructors. All computers in the lab are equipped with state-of-the-art 3D stereoscopic visualization hardware (3D LCD shutter glasses) and software.
Day One:
|
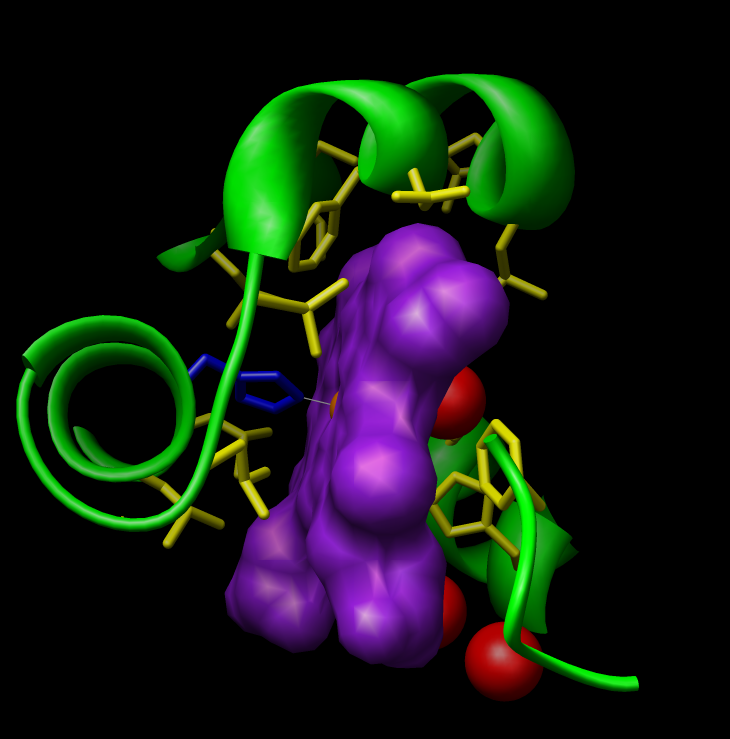
|
|
Day Two:
|
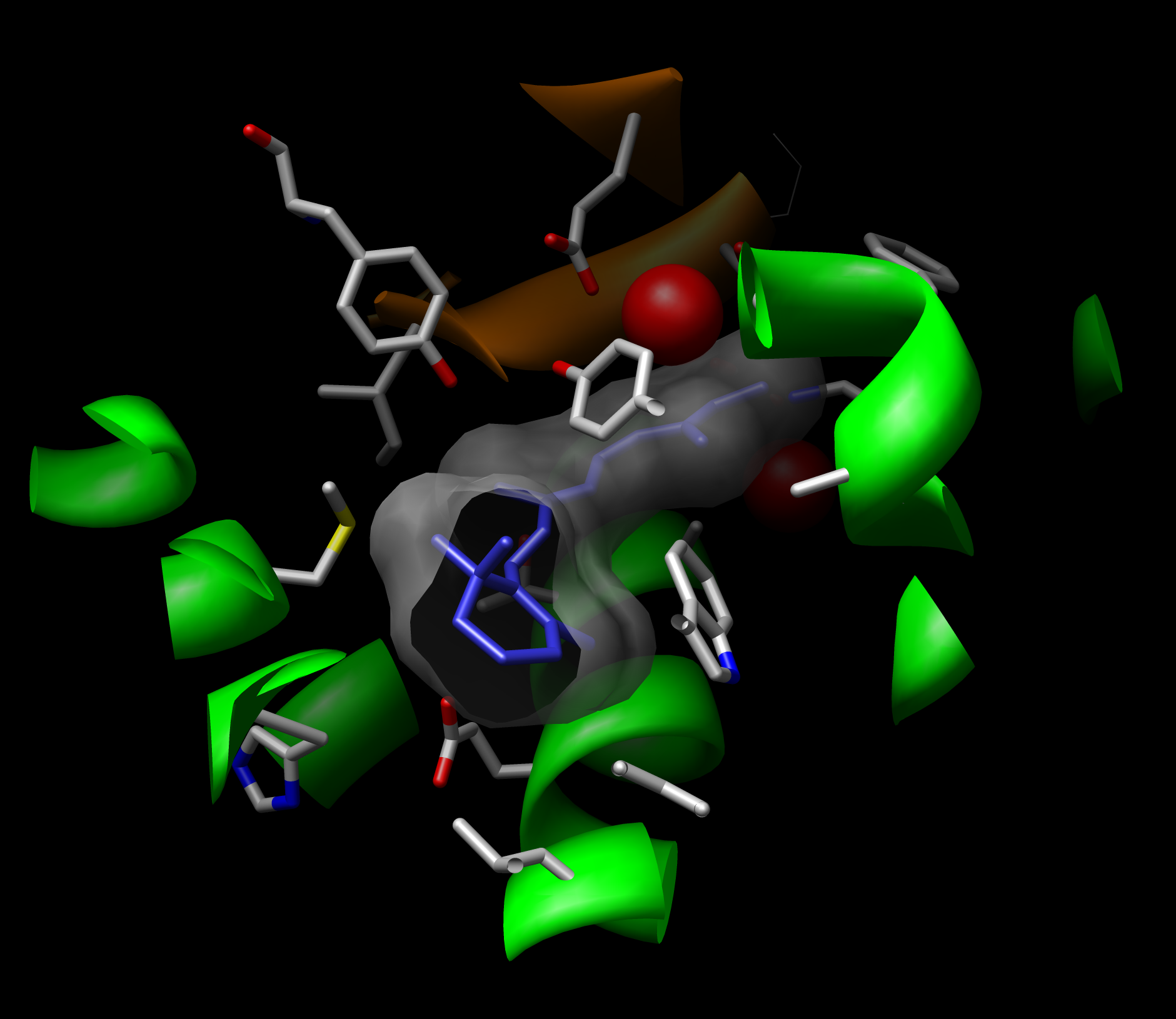
|
Interactive computer graphics techniques are frequently used to view and interrogate biomolecular structures. We will utilize USCF Chimera, a freely available molecular viewer for Microsoft Windows, Mac OSX, Linux, and UNIX-based computer operating systems.
You may wish to download Chimera and give it a try prior to the first workshop session. Chimera tutorials and program documentation are available online in the UCSF Chimera manual.
The Chimera Quick Reference Guide (pdf) is a convenient reference sheet for Chimera commands.
|
VU Home |
VUMC Home |
People Finder |
University Calendar
Jarrod Smith- modified on March 15, 2013 |