 | |||||||||||||||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
AMBER Archive (2009)
Subject: [AMBER] RE: question about TUTORIAL A1
From: Ross Walker (ross_at_rosswalker.co.uk)
Date: Mon Jul 06 2009 - 11:58:47 CDT
- Previous message: Ross Walker: "RE: [AMBER] Re: non aminoacid Residue"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
Hi Zeinab,
I reiterate my point that such messages should be sent to the AMBER mailing
list (amber_at_ambermd.org) instead of me directly.
Stop for a minute and think about some basic chemistry... How does a calcium
ion form 7 covalent bonds? Calcium is 1S2, 2S2, 2P6, 3S2, 3P6, 4S2 with the
2+ coming from the 4S2. Where does it get the electrons from to form 7
covalent bonds?
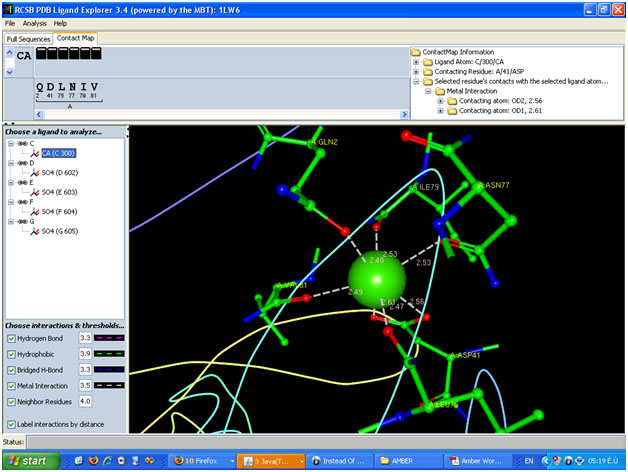
If you look at your image you will see that all of the interactions are
clearly ionic in nature. Thus as a first approximation you could probably
just model your calcium ion as a 2+ ionic charge. Run some test simulations
and see if it stays in place. If this is purely structural then you should
be okay.
If you wanted to take this further (as a hack for the limitations of a
classical model) then you might want to refit all of the nearby residues and
the Ca2+ charge, perhaps add some covalent bonds just to keep it in place.
Of course doing this you open a huge can of worms with respect to what is
actually physical, are the ion parameters for calcium actually any good in
the first place? You could probably do an entire Ph.D. on this question so
it is up to you whether you want to do this or not.
The short answer is that there is no definitive right or wrong way. This is
research! You just need to be able to make what you believe are justifiable
informed choices and then be able to defend these choices when it comes to
publishing your work.
Good luck,
Ross
From: zeinab bagheri [mailto:znbagheri_at_gmail.com]
Sent: Monday, July 06, 2009 6:36 AM
To: Ross Walker
Subject: Re: question about TUTORIAL A1
Dear Ross
thanks for your answer
A few questions. Are you sure this Calcium bonds to 7 atoms in amino acids?
This seems unlikely to me, are you sure it bonds to this much, with covalent
bonds? How are these formed?
yes I'm sure, I check in PDBsum home page and for confidence I check this
situation for other similar enzyme.
in the fallowing URLs , you can see LIGPLOT format of them:
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcod
e=1sup
<http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbco
de=1sup&template=ligands.html&o=METAL&l=1.1>
&template=ligands.html&o=METAL&l=1.1
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcod
e=1y1k
<http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbco
de=1y1k&template=ligands.html&o=METAL&l=1.1>
&template=ligands.html&o=METAL&l=1.1
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcod
e=1to2
<http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbco
de=1to2&template=ligands.html&o=METAL&l=1.1>
&template=ligands.html&o=METAL&l=1.1
furthermore I attached the image of the molecular fragment contain Ca++
(made by PDB ligand explorer )
I check the molecule in as the example in tutorial, I mean 1PLC , I think
the Cu atom bond to Met covalently and bond to the other atoms with metal
interaction . in my molecule Ca++ bond to all atom by metal interaction.
what should I do ?
I would start by looking in the literature to see if you can get parameters
for this. I would then as a first pass make this calcium a residue in its
own right with a charge of 2+.
excuse me , I don't understand . in the tutorial you say for driving the
partial charge , must use the RED server or REDIIIx program. is it correct?
another question? can I use antechamber for driving the charge instead of
using the RED server?
I think you mean; I don't need to change the place of Ca++ in the PDB's file
. is it correct?
if yes what should I do for the charge of the atoms near the Ca? I must
drive the charge of them with the method you said in the tutorial?
I would then build nonstandard versions of the residues that bond to this,
deleting the proton that is bound to the atom that bonds to the Ca and then
refit the charges on these amino acids.
OK. how I could delete the proton ? for example how can I delete the proton
of oxygen that bound the Ca++?
You can then bond these using the bond command in leap and provide
parameters through a frcmod file. This will do as a first pass, following on
from this you might want to consider a complete refit of the bound ligands
and the Ca atom together.
I'm very sorry for taking your time
Best Regards
ZnB
_______________________________________________
AMBER mailing list
AMBER_at_ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
- Previous message: Ross Walker: "RE: [AMBER] Re: non aminoacid Residue"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ]
|
VU Home |
VUMC Home |
People Finder |
University Calendar
webmaster- modified on July 06, 2009 |